The Methods we’re using…
Fly genetics
The fruit fly Drosophila melanogaster is very amenable to genetic modification. From the adult fly laying eggs to the 3rd instar larva we’re preferentially working with, it only takes 7-9 days. Using genetic toolboxes developed by other scientists, we can tag our protein of interest with fluorescent markers, or remove a protein the role of which we want to study.
Since its initial use by William Castles and Thomas Hunt Morgan more than 100 years ago, Drosophila melanogaster has become one of the most heavily used model organisms to study a diverse range of biological processes including genetics, embryonic development, learning, aging, and neurobiological processes. Its initial use by Morgan redefined the theory of heredity by discovering genes and found that they are located within chromosomes. This led to the first of six Nobel prizes awarded for Drosophila research (the others were awarded for the discovery that mutations are caused by X-ray, for founding the field of evolutionary developmental biology, and for the discovery of olfactory receptors, the innate immunity and finally of the circadian rhythm).
Interestingly, one convention unique to Drosophila was introduced by Morgan: the naming of genes in accordance to the phenotype their knock-down causes: the first mutation ever isolated is known as white, causing white instead of the wild type red eyes. Over the years, this convention has also led to somewhat entertaining names as “cheap-date”, naming flies that suffer from a reduced alcohol tolerance; or the opposite situation “happy hour”, naming flies that show an increased tolerance (Corl et al., 2009; Moore et al., 1998). For the research questions our lab is focused on, Drosophila is unique in its advantages. These include a relatively short generation time of about 10 days (see life cycle), an every growing genetic tool box allowing tissue- and even cell-specific expression or knock-down of genes of interest but especially a tremendous applicability to scientific methodologies (see methods section). Furthermore, 75% of all known human disease related genes have a recognizable counterpart in the Drosophila genome (Jennings, 2011) making it a perfectly suited tool to not just understand the molecular underpinnings of humans diseases, but also to study the function of these molecules in the healthy organism.
Live Imaging
Biological processes are best viewed in real time and with as much temporal and spatial information as possible. Therefore, we use fluorescent probes to visualize synaptic activity and protein trafficking in live dissected larvae.
The fluorescent calcium indicator GCaMP has proven especially useful in this, and we use it to watch as the synapse releases its vesicles. Using this method, we measure the influence of synaptic proteins on signal transmission which helps us understand how the synapse works.
Super-resolution microscopy: STED
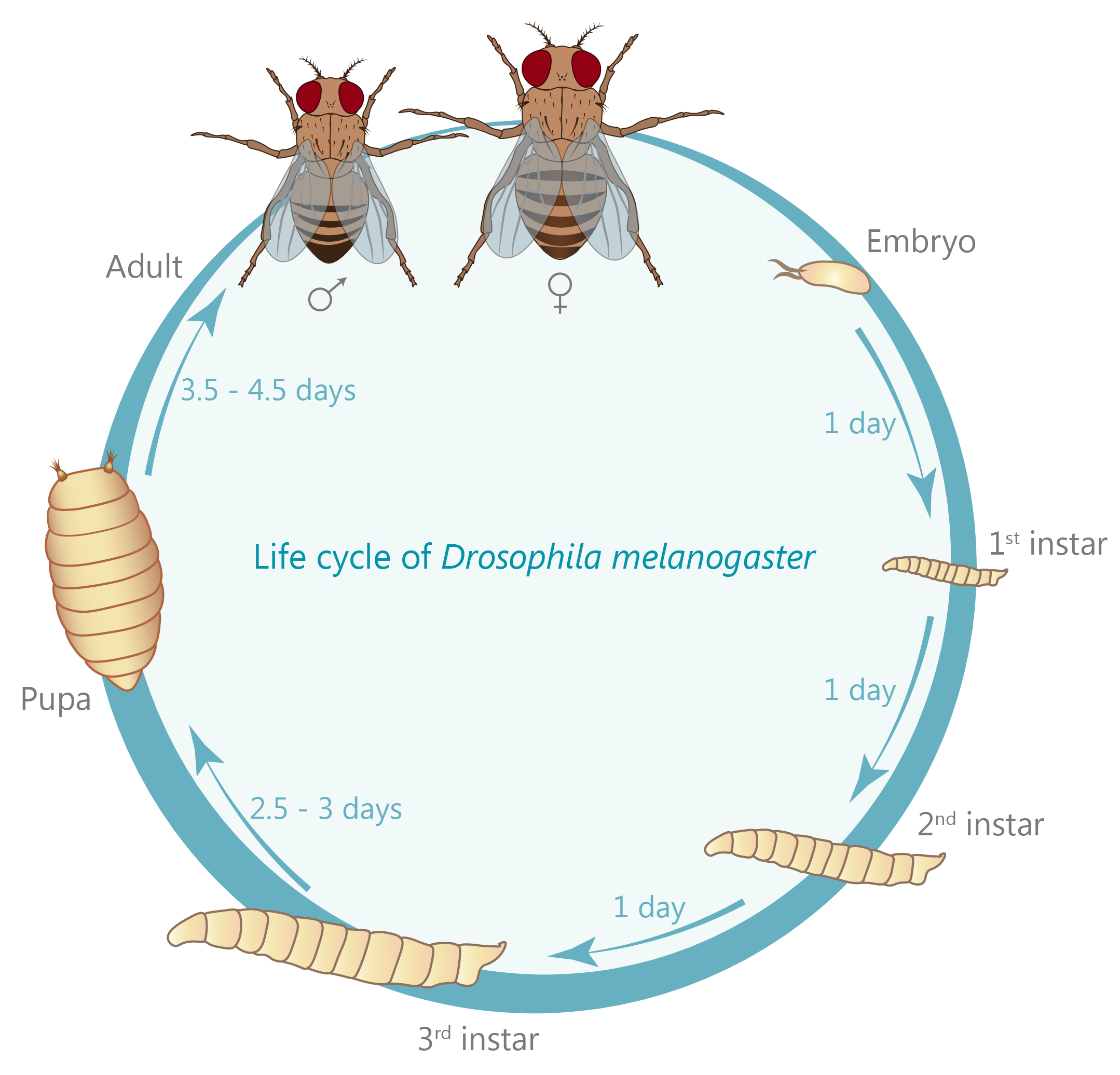
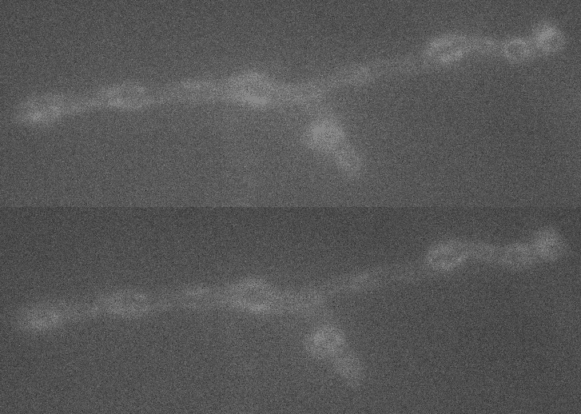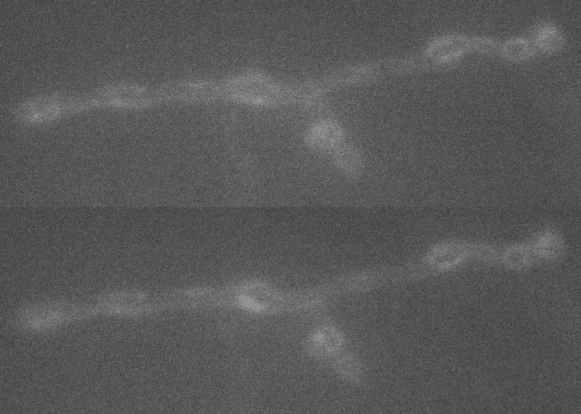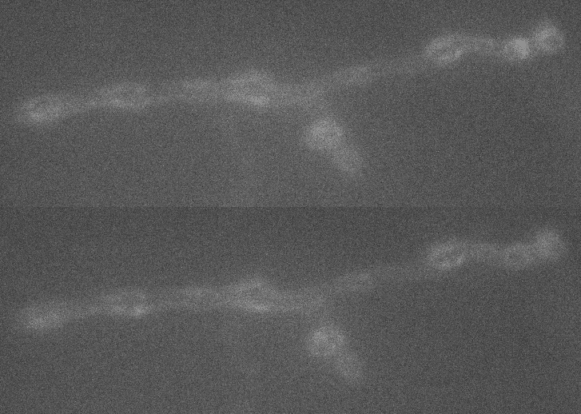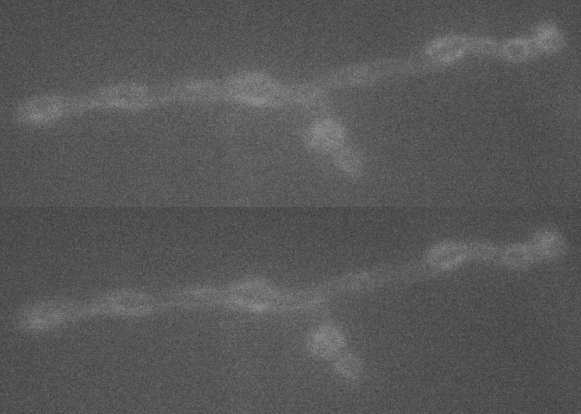
A large extent of the current understanding of biological processes comes from experiments that visualized them using various microscopic techniques including fluorescence microscopy. However, due to the Abbe diffraction limit of light conventional fluorescence microscopes are limited in their resolution to ~250 nm (approximately equal to half the wavelength of the used light beam). Nevertheless, cellular structures including synapses but especially the proteins they are composed of are far smaller, partially by more than a factor of 10. As a conventional fluorescent microscope illuminates all fluorophores within a spot at the same time, they also emit light at the same time. Therefore, two points in closer proximity than the full-width-half-maximum (FWHM) of their point spread function (PSF) will be difficult to resolve, as both PSF overlap to a certain degree. The invention of super-resolution microscopy broke the diffraction barrier within the last 20 years and opened a complete new field of science. In our lab we mainly use the super-resolution technique Stimulated Emission Depletion microscopy (STED).
For STED microscopy, two overlapping, synchronized laser beams are used, which arrive at the same sample position consecutively whereas the first laser excites the fluorophore (excitation laser) and forces an electron to a higher energy level whereas the second one (STED or depletion laser) de-excites/depletes it and forces the electron back to the ground state. In the region where both lasers are present, stimulated depletion is happening, while in a focal spot where just the excitations laser is present, spontaneous emission and therefore fluorescence is observed.
Electrophysiology
We use the Drosophila larval neuromuscular junction as a model to investigate neurotransmission. Here motoneurons innervate muscle cells forming a large number of synaptic connections. We insert sharp glass electrodes into larval muscles and measure the electrical properties of the membrane by making use of two different techniques:
Current clamp records changes in the membrane potential. Two electrode voltage clamp records the current injected to maintain a fixed holding potential of the muscle membrane. We investigate evoked neurotransmission by stimulating the presynaptic axon as well as spontaneous activity. We avail of different pharmacological agents to identify underlying mechanisms of synaptic function.
Computational Biology &
Mathematical Modelling
Experimental Data is not always sufficient to fully understand the biological processes under investigation. Therefore, we employ computer-aided methods that help us build simulations and models of the synapse. Making assumptions about what processes are involved in a synaptic process, we formulate mathematical equations and have the computer solve them.
The simulations we derive from this can then be compared to experimental data. If our simulations do not agree with the experimental data, we know that we are making the wrong assumptions and can adapt the model until we find which underlying process we are missing.